Introduction
Last updated on 2025-09-30 | Edit this page
Overview
Questions
- What are expression quantitative trait loci (eQTL)?
- How are eQTL used in genetic studies?
Objectives
- Describe how an expression quantitative trait locus (eQTL) impacts gene expression.
- Describe how eQTL are used in genetic studies.
Introduction
Differences in disease risk between individuals are often caused by genetic variants. Identifying the effects of genetic variants is key to understanding disease phenotypes and their underlying biology. The effects of variants in many single-gene disorders, such as cystic fibrosis, are generally well-characterized and their disease biology well understood. For example, in cystic fibrosis, mutations in the coding region of the CFTR gene alter the three-dimensional structure of the chloride channel proteins in epithelial cells, affecting not only chloride transport, but also sodium and potassium transport in the lungs, pancreas and skin. The path from gene mutation to altered protein to disease phenotype is relatively simple and well understood.
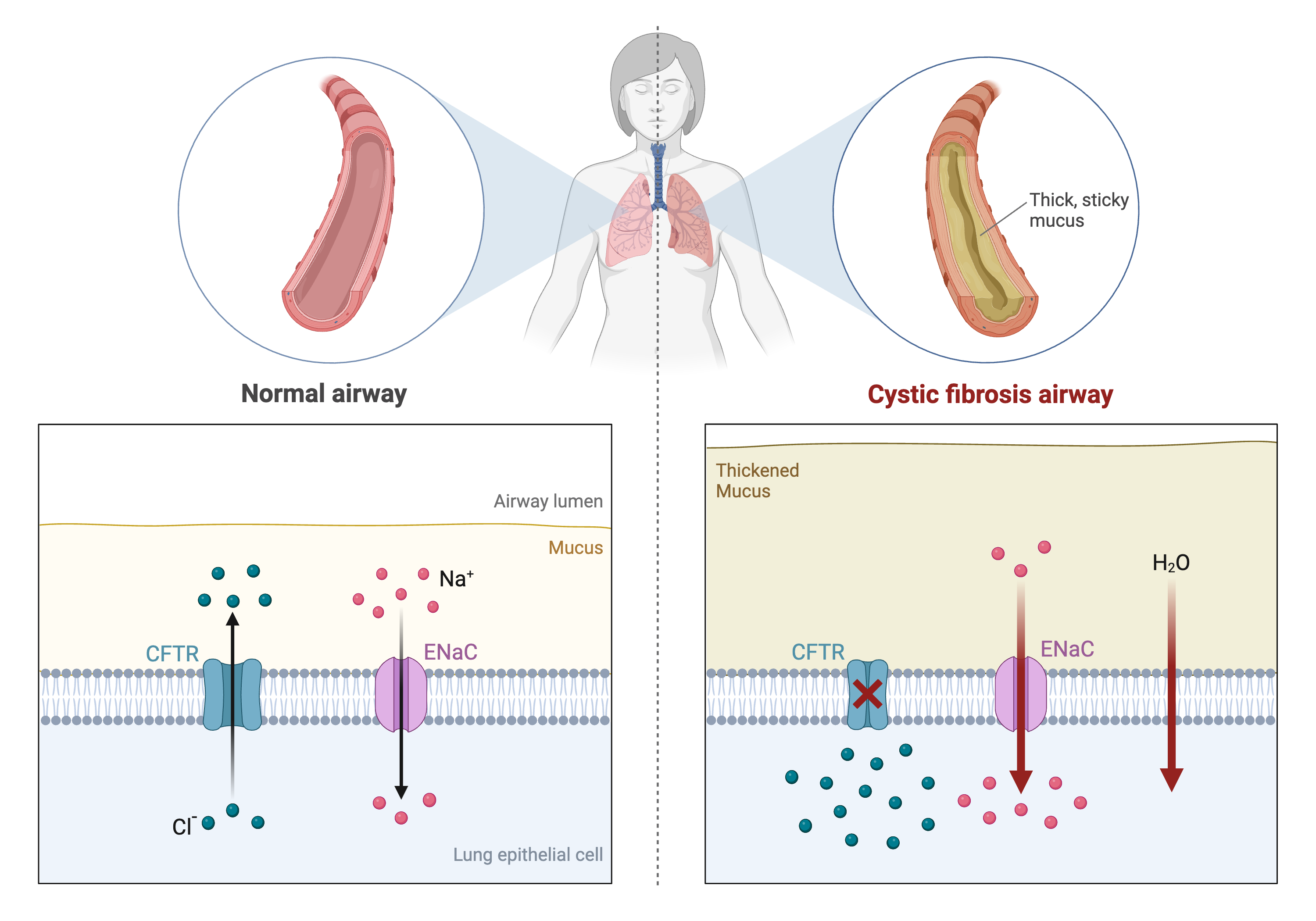
Created in BioRender
The most common human disorders, however, involve many genes interacting with each other and with the environment, a far more complicated path to follow than the path from a single gene mutation to its protein to a disease phenotype. Cardiovascular disease, Alzheimer’s disease, arthritis, diabetes and cancer involve a complex interplay of genes with environment, and their mechanisms are not well understood. One method of understanding the relationship between genetic variants and disease is a genome-wide association study (GWAS), which associates genetic variants with disease traits. It is tempting to think that these genetic variants would fall in coding regions. However, most GWAS variants for common diseases like diabetes are located in non-coding regions of the genome. These variants are therefore likely to fall in regulatory sequences which are involved in gene regulation.
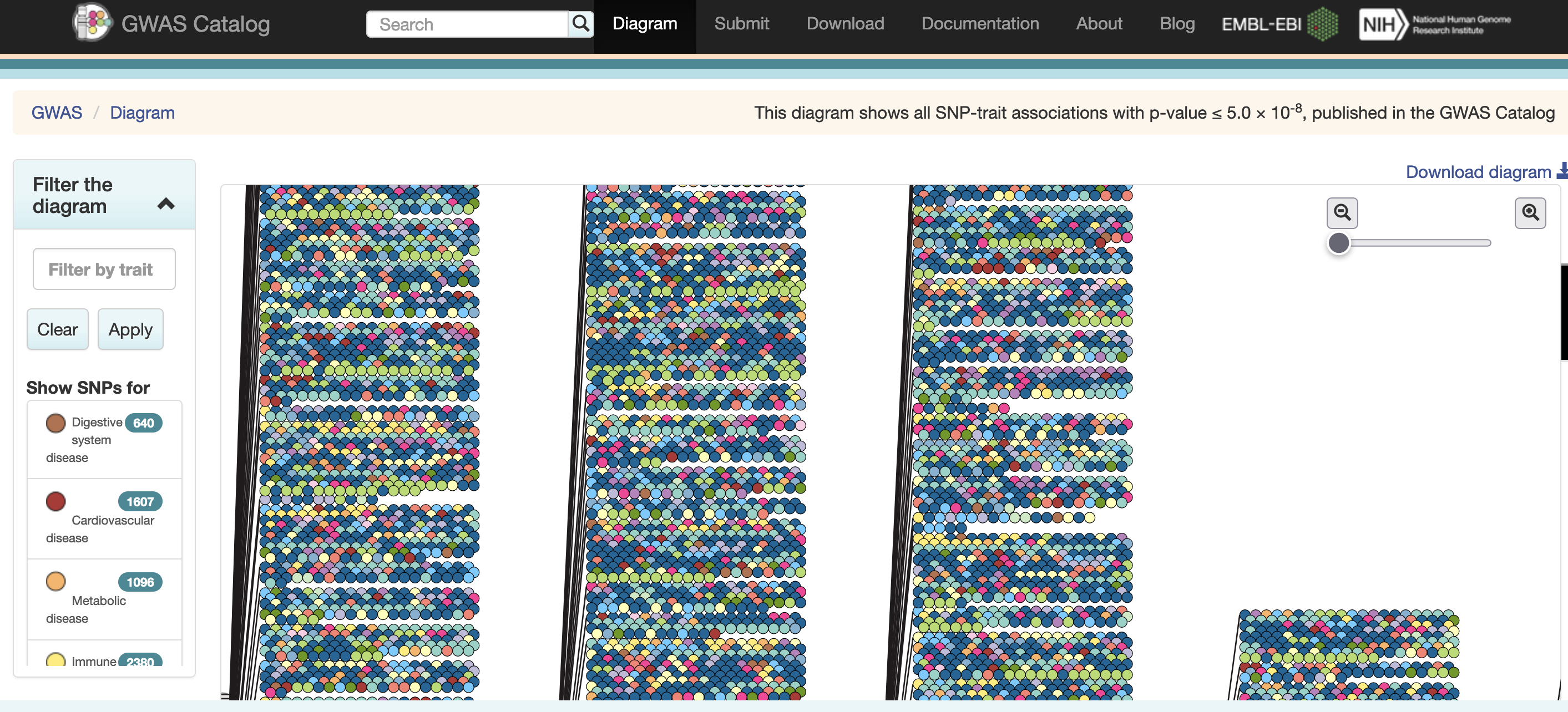 Excerpted from the GWAS Catalog
Excerpted from the GWAS Catalog
Gene regulation controls the quantity, timing and locale of gene expression. Analyzing the association between gene expression and genetic variants is known as expression quantitative trait locus (eQTL) mapping.
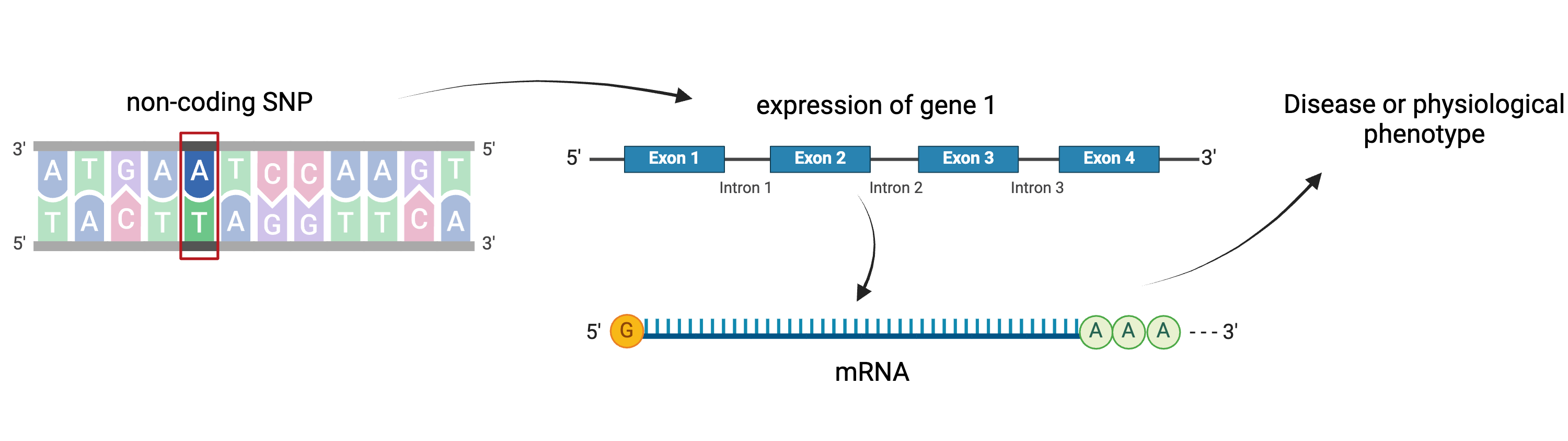 Created
in BioRender
Created
in BioRender
eQTL mapping searches for associations between the expression of one or more genes and a genetic locus. Specifically, genetic variants underlying eQTL peaks explain some of the variation in gene expression levels. eQTL studies can reveal the architecture of quantitative traits, connect DNA sequence variation to phenotypic variation, and shed light on transcriptional regulation and regulatory variation. Traditional analytic techniques like linkage and association mapping can be applied to thousands of gene expression traits (transcripts) in eQTL analysis, such that gene expression can be mapped in much the same way as a physiological phenotype like blood pressure or heart rate. Joining gene expression and physiological phenotypes with genetic variation can identify genes with variants affecting disease phenotypes.
To the simple diagram above we’ll add two more details. Non-coding SNPs can regulate gene expression from nearby locations on the same chromosome:
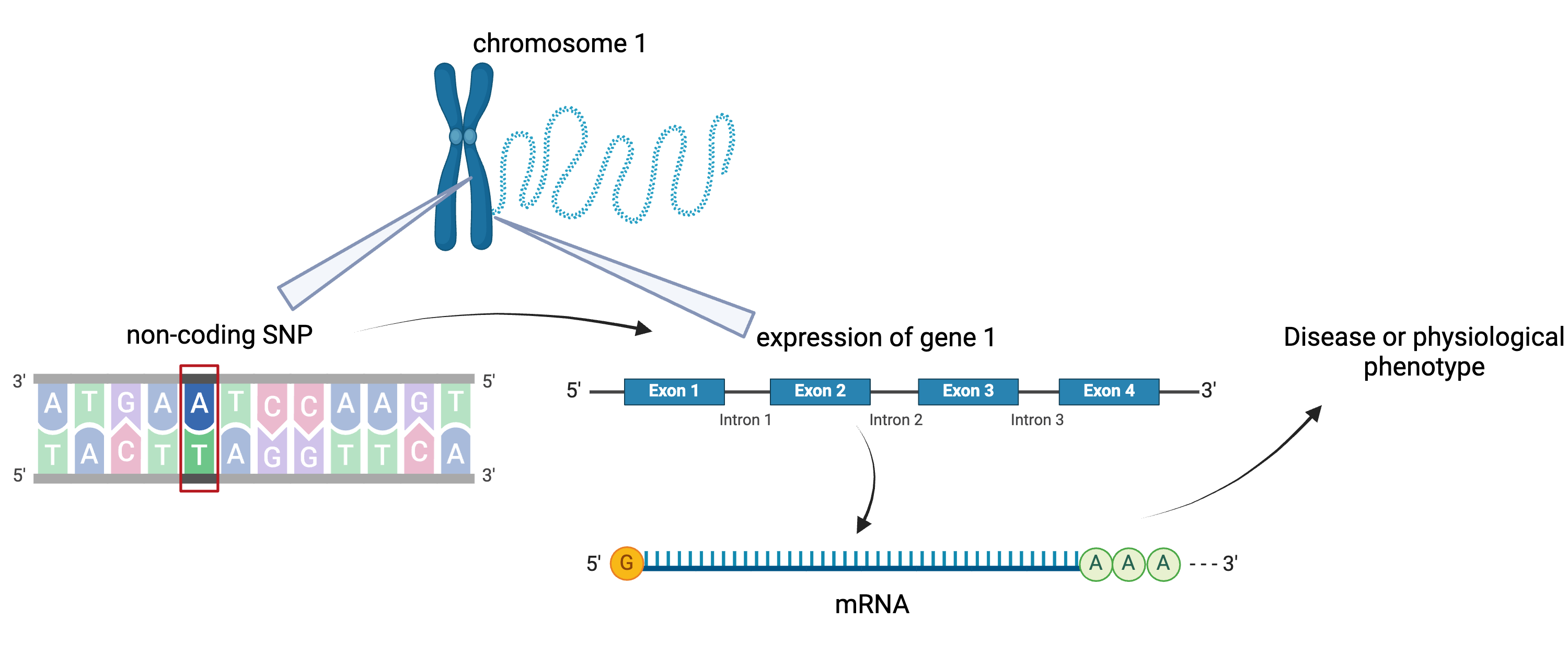 Created in BioRender
Created in BioRender
SNPs that affect gene expression from afar, often from a different chromosome from the gene that they regulate are called distal regulators.
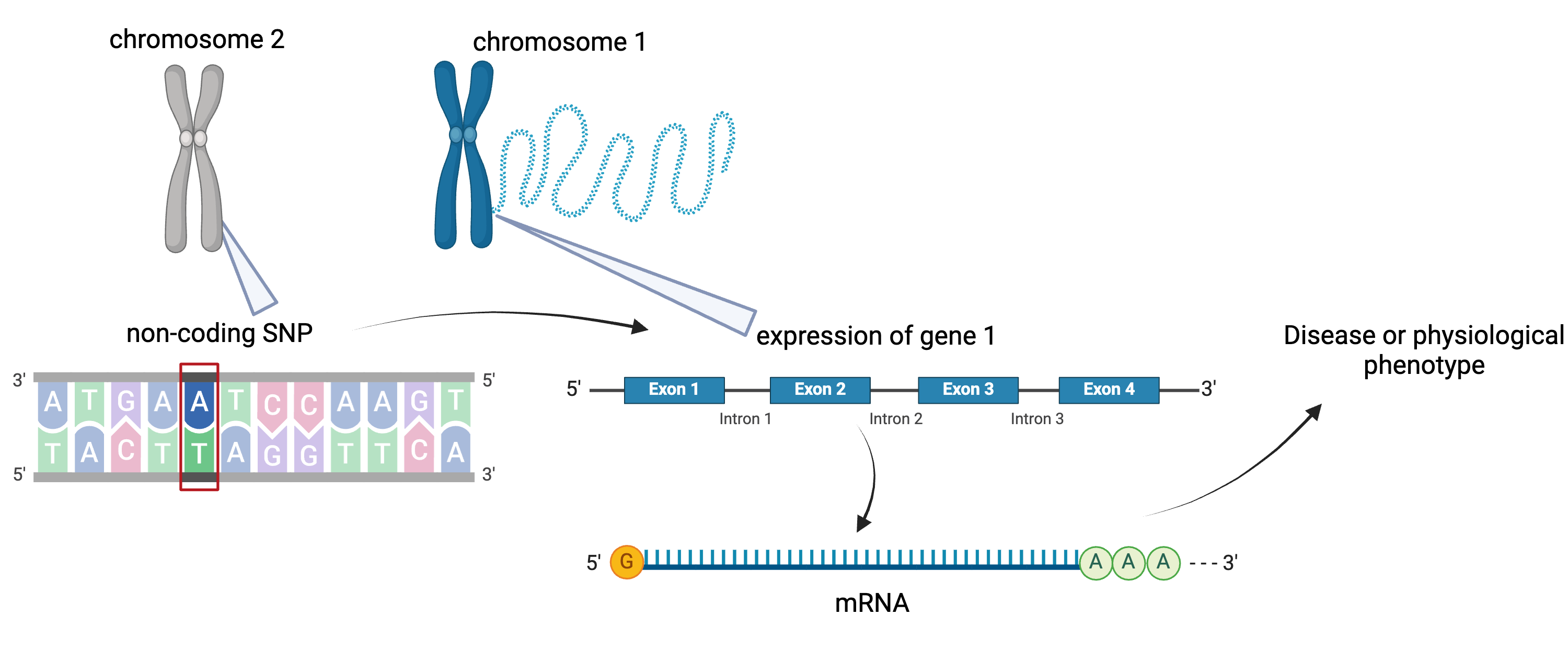
Created in BioRender
- An expression quantitative trait locus (eQTL) explains part of the variation in gene expression.
- Traditional linkage and association mapping can be applied to gene expression traits (transcripts).
- Genetic variants, such as single nucleotide polymorphisms (SNPs), that underlie eQTL illuminate transcriptional regulation and variation.
