Factors
Overview
Teaching: 25 min
Exercises: 20 minQuestions
How do I work with factors in R?
Objectives
Describe what a factor is.
Convert between strings and factors.
Reorder and rename factors.
Factors in R
When we did str(tumor) we saw that most of the columns are numeric.
The column Grp, however, isof the class character.
This column contains categorical data, that is, it can only take on
a limited number of values. To see this, use the unique() function.
unique(tumor$Grp)
[1] "1.CTR" "2.D" "3.R" "4.D+R"
As a reminder:
Cells from a human glioma cell line were implanted in the flank of n=37 nude mice and a subcutaneous tumor (xenograft) was allowed to grow. When a tumor grew to around 40-60 $mm^3$, the animal was assigned to one of 4 experimental groups (day 0):
1) Control (CTR, n=8);
2) Drug only (D, n=10);
3) Radiation only (R, n=10); and
4) Drug + Radiation (D+R, n=9).
The main outcome in xenograft experiments is the size (volume) of the tumor over time. The study’s two main scientific aims were to assess whether:
a. The drug has an effect on tumor growth.
b. The administration of the drug before radiation enhances the effect of the latter on tumor growth.
R has a special class for working with categorical data, called factor.
Factors are very useful and actually contribute to making R particularly well
suited to working with data. So we are going to spend a little time introducing
them.
Once created, factors can only contain a pre-defined set of values, known as levels. Factors are stored as integers associated with labels and they can be ordered or unordered. While factors look (and often behave) like character vectors, they are actually treated as integer vectors by R. So you need to be very careful when treating them as strings.
When importing a data frame with read_csv(), the columns that contain text are not automatically coerced (=converted) into the factor data type, but once we have loaded the data we can do the conversion using the factor() function:
tumor$Grp <- factor(tumor$Grp)
We can see that the conversion has worked by using the summary()
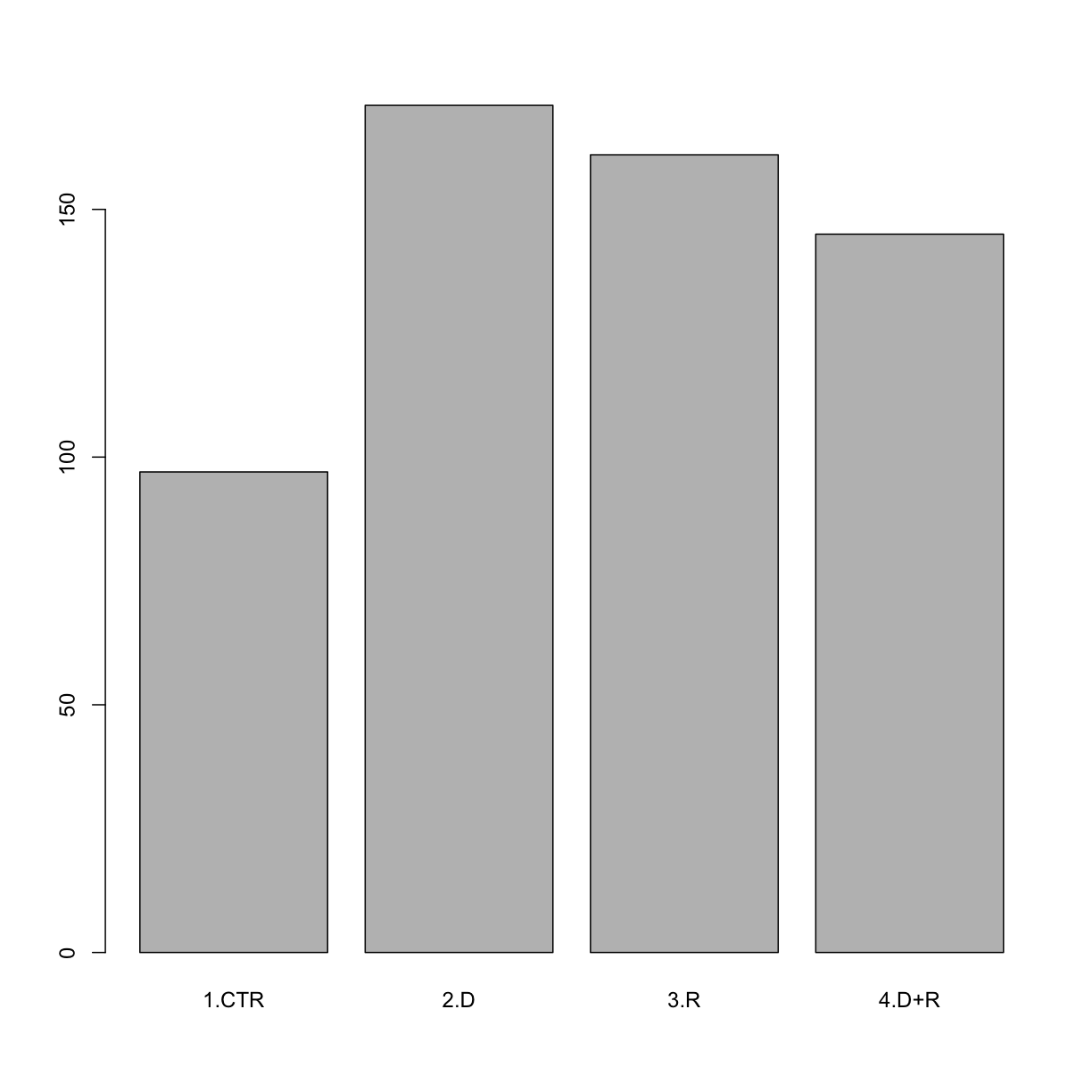
function again. This produces a table with the counts for each factor level:
summary(tumor$Grp)
1.CTR 2.D 3.R 4.D+R
97 171 161 145
By default, R always sorts levels in alphabetical order. For instance, if you have a factor with 2 levels:
direction <- factor(c("right", "left", "left", "right"))
R will assign 1 to the level "left" and 2 to the level "right" (because
l comes before r, even though the first element in this vector is
"right"). You can see this by using the function levels() and you can find the
number of levels using nlevels():
levels(direction)
[1] "left" "right"
nlevels(direction)
[1] 2
Sometimes, the order of the factors does not matter, other times you might want
to specify the order because it is meaningful (e.g., “low”, “medium”, “high”),
it improves your visualization, or it is required by a particular type of
analysis. Here, one way to reorder our levels in the direction vector would be:
direction # current order
[1] right left left right
Levels: left right
direction <- factor(direction, levels = c("right", "left"))
direction # after re-ordering
[1] right left left right
Levels: right left
In R’s memory, these factors are represented by integers (1, 2), but are more
informative than integers because factors are self describing: "left",
"right" is more descriptive than 1, 2. Which one is “right”? You wouldn’t
be able to tell just from the integer data. Factors, on the other hand, have
this information built in. It is particularly helpful when there are many levels
(like the Grp names in our example dataset).
Exercise
- Change the column
Grpin thetumordata frame into a factor.- Using the functions you learned before, can you find out:
- How many were in group D?
- How many levels there are?
Solution
tumor$Grp <- factor(tumor$Grp) summary(tumor$Grp)1.CTR 2.D 3.R 4.D+R 97 171 161 145nlevels(tumor$Grp)[1] 4
Converting factors
If you need to convert a factor to a character vector, you use
as.character(x).
as.character(direction)
[1] "right" "left" "left" "right"
In some cases, you may have to convert factors where the levels appear as
numbers (such as concentration levels or years) to a numeric vector. For
instance, in one part of your analysis the years might need to be encoded as
factors (e.g., comparing average weights across years) but in another part of
your analysis they may need to be stored as numeric values (e.g., doing math
operations on the years). This conversion from factor to numeric is a little
trickier. The as.numeric() function returns the index values of the factor,
not its levels, so it will result in an entirely new (and unwanted in this case)
set of numbers. One method to avoid this is to convert factors to characters,
and then to numbers.
Another method is to use the levels() function. Compare:
year_fct <- factor(c(1990, 1983, 1977, 1998, 1990))
as.numeric(year_fct) # Wrong! And there is no warning...
[1] 3 2 1 4 3
as.numeric(as.character(year_fct)) # Works...
[1] 1990 1983 1977 1998 1990
as.numeric(levels(year_fct))[year_fct] # The recommended way.
[1] 1990 1983 1977 1998 1990
Notice that in the levels() approach, three important steps occur:
- We obtain all the factor levels using
levels(year_fct) - We convert these levels to numeric values using
as.numeric(levels(year_fct)) - We then access these numeric values using the underlying integers of the
vector
year_fctinside the square brackets
Renaming factors
When your data is stored as a factor, you can use the plot() function to get a
quick glance at the number of observations represented by each factor
level. Let’s look at the number in each group.
## bar plot of the numbers in each group:
plot(tumor$Grp)
Exercise
We have seen how data frames are created when using
read_csv(), but they can also be created by hand with thedata.frame()function. There are a few mistakes in this hand-crafteddata.frame. Can you spot and fix them? Don’t hesitate to experiment!animal_data <- data.frame( animal = c(dog, cat, sea cucumber, sea urchin), feel = c("furry", "squishy", "spiny"), weight = c(45, 8 1.1, 0.8) )Can you predict the class for each of the columns in the following example? Check your guesses using
str(country_climate):
- Are they what you expected? Why? Why not?
- What would you need to change to ensure that each column had the accurate data type?
country_climate <- data.frame( country = c("Canada", "Panama", "South Africa", "Australia"), climate = c("cold", "hot", "temperate", "hot/temperate"), temperature = c(10, 30, 18, "15"), northern_hemisphere = c(TRUE, TRUE, FALSE, "FALSE"), has_kangaroo = c(FALSE, FALSE, FALSE, 1) )Solution
- missing quotations around the names of the animals
* missing one entry in thefeelcolumn (probably for one of the furry animals)
* missing one comma in theweightcolumncountry,climate,temperature, andnorthern_hemisphereare characters;has_kangaroois numeric
- using
factor()one could replace character columns with factors columns- removing the quotes in
temperatureandnorthern_hemisphereand replacing 1 by TRUE in thehas_kangaroocolumn would give what was probably intended
The automatic conversion of data type is sometimes a blessing, sometimes an annoyance. Be aware that it exists, learn the rules, and double check that data you import in R are of the correct type within your data frame. If not, use it to your advantage to detect mistakes that might have been introduced during data entry (for instance, a letter in a column that should only contain numbers).
Learn more in this RStudio tutorial
Key Points