Input File Format
Last updated on 2025-09-16 | Edit this page
Overview
Questions
- How are the data files formatted for qtl2?
- Which data files are required for qtl2?
- Where can I find sample data for mapping with the qtl2 package?
Objectives
- To specify which input files are required for qtl2 and how they should be formatted.
- To locate sample data for qtl mapping.
QTL mapping data consists of a set of tables of data: sample genotypes, phenotypes, marker maps, etc. These different tables are in different comma-separated value (CSV) files. In each file, the first column is a set of IDs for the rows, and the first row is a set of IDs for the columns. For example, the genotype data file will have individual IDs in the first column, marker names for the rest of the column headers.
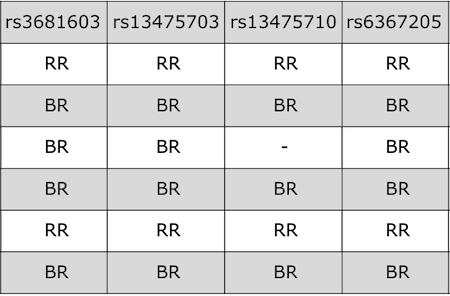
The sample genotype file above shows two alleles: B and R. These represent the founder strains for an intercross, which are C57BL/6 (BB) and BTBR (RR) Tian et al. The B and R alleles themselves represent the haplotypes inherited from the parental strains C57BL/6 and BTBR.
For the purposes of learning QTL mapping, this lesson begins with an
intercross that has only 3 possible genotypes instead of 8 or 36. Once
we have learned how to use qtl2 for the simpler case, we
will advance to the most complex case involving mapping in DO mice.
R/qtl2 accepts the following files:
1. genotypes
2. phenotypes
3. phenotype covariates (i.e. tissue type, time points)
4. genetic map
5. physical map (optional)
6. control file (YAML or JSON format, not CSV).
We use both a genetic marker map and a physical map (if available). A sample from a genetic map of SNP markers is shown here.

A physical marker map provides location in bases rather than centiMorgans.

Numeric phenotypes are separate from the often non-numeric covariates.

Phenotype covariates are metadata describing the phenotypes. For example, in the case of a phenotype measured over time, one column in the phenotype covariate data could be the time of measurement. For gene expression data, we would have columns representing chromosome and physical position of genes, as well as gene IDs. The covariates shown below include sex and parental grandmother (pgm).

In addition to the set of CSV files with the primary data, we need a separate control file with various control parameters (or metadata), including the names of all of the other data files and the genotype codes used in the genotype data file. The control file is in a specific format using either YAML or JSON; these are human-readable text files for representing relatively complex data.
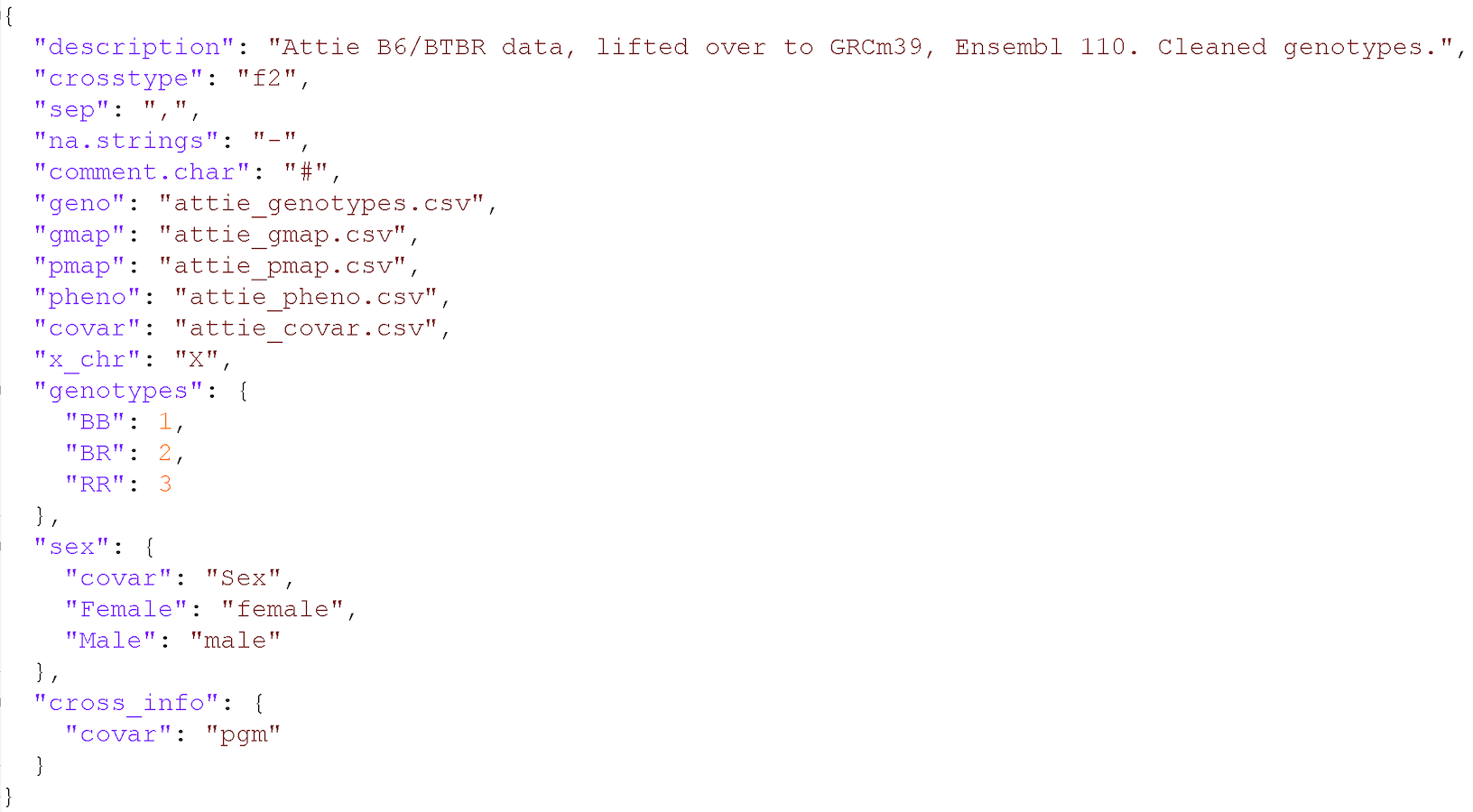
A big advantage of this control file scheme is that it greatly
simplifies the function for reading in the data. That function,
read_cross2(), has a single argument: the name
(with path) of the control file.
For further details, see the separate vignette on the input file format.
Challenge 1: What data does qtl2 need?
- Which data files are required by
qtl2? - Which ones are optional?
- How should they be formatted?
- genotypes, phenotypes, genetic map
- physical map
- csv; JSON or YAML for control file
Sample data sets
In this lesson, we will not work with data sets included in the
qtl2 package, though you may want to explore them to learn
more. You can find out more about the sample data
files from the R/qtl2 web site. Zipped versions of these datasets
are included with the qtl2geno package and can be
loaded into R using the read_cross2() function. Additional
sample data sets, including data on Diversity Outbred (DO) mice, are
available at https://github.com/rqtl/qtl2data.
Challenge 2: Additional R/qtl2 datasets
Go to https://github.com/rqtl/qtl2data to view additional
sample data.
1). Find the Recla data and locate the phenotype data file. Open the
file by clicking on the file name. What is in the first column? the
first row?
2). Locate the genotype data file, click on the file name, and view the
raw data. What is in the first column? the first row?
3). Locate the covariates file and open it by clicking on the file name.
What kind of information does this file contain?
4). Locate the control file (YAML or JSON format) and open it. What kind
of information does this file contain?
1). What is in the first column of the phenotype file? Animal ID. The
first row? Phenotype variable names - OF_distance_first4, OF_distance,
OF_corner_pct, OF_periphery_pct, …
2). What is in the first column of the genotype file? marker ID. the
first row? Animal ID - 1,4,5,6,7,8,9,10, …
3). Locate the covariates file and open it. What kind of information
does this file contain? Animal ID, sex, cohort, group, subgroup, ngen,
and coat color.
4). Locate the control file (YAML or JSON format) and open it. What kind
of information does this file contain? Names of primary data files,
genotype and allele codes, cross type, description, and other
metadata.
- QTL mapping data consists of a set of tables of data: genotypes, phenotypes, marker maps, etc.
- These different tables are in separate comma-delimited (CSV) files.
- In each file, the first column is a set of IDs for the rows, and the first row is a set of IDs for the columns.
- In addition to primary data, a separate file with control parameters (or metadata) in either YAML or JSON format is required.
- Published and public data already formatted for QTL mapping are available on the web.
- These data can be used as a model for formatting your own QTL data.
